Imaging Neurotransmitters with AAV GRAB Sensors

The complex function in the brain is inseparable from the efficient and specific information exchange and integration between neurons. Neurotransmitters (such as dopamine, acetylcholine, and norepinephrine) are key biological small molecules and participate in the process of information transmission mediated by chemical synapses between neurons and are involved in a variety of physiological functions including development, information perception, learning/memory, emotion, motor regulation and higher-level cognitive behavior in the brain. Malfunction of neurotransmitters and receptors will cause devastating diseases, such as Parkinson's disease, Alzheimer's disease, addiction, and epilepsy. Therefore, the study of neurotransmitters is essential for understanding the mechanism of brain function.
Existing Tools for Neurotransmitter Imaging & Their Limitations
However, precise measurements of the spatial and temporal patterns of release of neurotransmitters, specifically dopamine (DA), during complex behaviors are lacking, due in large part to the limitations of existing methods for DA detection. Intracerebral microdialysis has long been the gold standard for quantitative measurements of extracellular DA concentration. However, its relatively slow sampling rate (> 1 min between sampling, typically ~10 mins) (Chefer et al., 2009) is not well suited to detect DA dynamic changes during complex and rapidly evolving behaviors.
Fast-scan cyclic voltammetry (FSCV) is an electrochemical method that can measure changes in extracellular DA concentrations with 10 ms temporal resolution and ~1 nM sensitivity (Robinson et al., 2008). However, FSCV requires substrate oxidization for signal detection, therefore it is not readily able to distinguish DA from other structurally similar neurotransmitters, such as norepinephrine (NE) (Robinson et al., 2003). Moreover, both microdialysis and FSCV require implantation of a relatively large probe (approximately 70–300 μm in diameter) into brain tissue, which limits the ability to achieve spatially precise measurements of endogenous DA release (Jaquins-Gerstl and Michael, 2015).
The development of a genetically encoded fluorescent sensors, which we call GPCR Activation-based (GRAB) sensors, has overcome the limitations of past methods and has enabled direct, rapid, sensitive, and cell-type specific detection of extracellular neurotransmitters. GRAB neurotransmitter sensors were initially developed by Dr. Yulong Li’s Lab at Peking University.
The sensors were engineered by coupling a conformationally sensitive circular-permutated EGFP (cpEGFP) to a selected human neurotransmitter receptor, such as DA receptor. For instance, GRABDA sensors were constructed by replacing the third intracellular loop (IL3) of the human Dopamine D1 (DRD1), D2 (DRD2) and D4 receptors (DRD4) with a cpGFP module from GCaMP6 (Patriarchi et al., 2018). Upon binding of neurotransmitters, such as dopamine, to the engineered receptor, a ligand-stabilized conformational change occurs in the receptor, which in turn results in an optical response of the conformation-sensitive fluorescent protein inserted in the receptor’s third intracellular loop (Sun et al., 2020). Therefore, changes in the fluorescence intensity of fluorescent proteins can be used to indicate dynamic changes in the concentration of neurotransmitters. These sensors enable real-time detection of endogenous neurotransmitters in acute brain slices of mice and in the intact brains of versatile animal models including flies, fish, and mice.
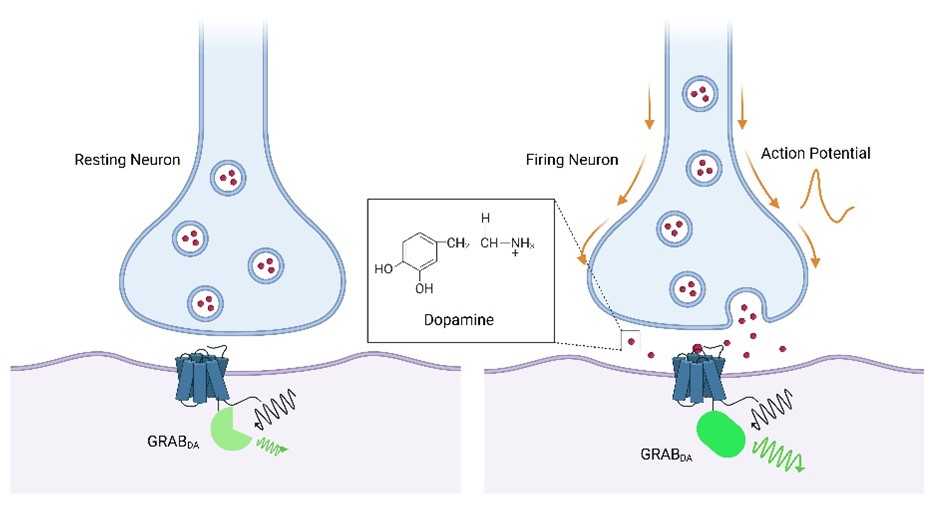
Figure 1. Schematic diagram showing the working principle of GRABDA Sensor. Upon binding of dopamine to the engineered receptor, a ligand-stabilized conformational change occurs in the receptor, which in turn results in an optical response of the conformation-sensitive fluorescent protein inserted in the receptor’s third intracellular loop.
Advantages of the neurotransmitter GRAB probe:
- 1) High sensitivity;
- 2) High specificity;
- 3) High temporal and spatial resolution;
- 4) High signal-to-noise ratio;
- 5) Non-invasive;
- 6) No need to build additional detection equipment, it is consistent with the calcium signal detection system.
At present, more than 10 new GRAB probes (including dopamine, acetylcholine, norepinephrine, etc.) have been published in relevant literature. Researchers can express probes in a variety of cultured cells, mouse brain slices, or living Drosophila, zebrafish, mice and other model animals through transfection, virus injection, and construction of transgenic animals.
| Name | Function |
|---|---|
| DA/dLight | Dopamine probe |
| Ach | Acetylcholine probe |
| NE | Norepinephrine probe |
| SHT/5-HT/ iSeroSnFR | Serotonin probe |
| Ado | Adenosine probe |
| ATP/ iATPSnFRs | Adenosine triphosphate probe |
| CCK | Cholecystokinin probe |
| VIP | Vasoactive intestinal peptide probe |
| eCB | Endocannabinoid probe |
| iGABASnFR | GABA probe |
| iGluSnFR | Glutamate probe |
| ArcLight | Voltage indicator |
| ASAP | Voltage indicator |
| CRF | Corticotropin-releasing factor probe |
| SST | Somatostatin probe |
| OXT | Oxytocin probe |
| OxLight | Orexins probe |
| HA | Histamine Probe |
| iGlucoSnFR | Glucose probe |
Fluorescence Selection
Red fluorescent sensors have distinct and well-separated spectra from those of green fluorescent protein (GFP)–based sensors and blue light–excitable channelrhodopsin 2 (ChR2), thus enabling the orthogonal readout of distinct neurochemical events, or simultaneous monitoring of neurotransmitter release and blue light–mediated control of neuronal activity. Moreover, red fluorescent sensors require excitation with a relatively longer wavelength, providing additional advantages over GFP, including reduced phototoxicity, reduced background, and deeper tissue penetration (Shu et al, 2009; Alford et al., 2013). Starting with the dopamine D2 receptor (D2R) and the conformation-sensitive red fluorescent protein cpmApple18, Dr. Yulong Li’s lab at Peking University has generated red fluorescent DA sensors called rGRABDA1m and rGRABDA1h (abbreviated rDA1m and rDA1h, respectively), with a dynamic range similar to the corresponding first-generation green DA sensors.
All GRAB sensors (including the latest ACh, DA, NE, 5-HT, Ado, ATP, VIP, CCK and eCB sensors) with AAV are available. To view the probe list, please click here or contact orders@biohippo.com. Please email orders@biohippo.com if the AAV you are looking for is not listed. We can design and package the AAV vectors to serve your special research needs.
FAQs
A1: h/m/l are the abbreviations for high/median/low, which indicates the sensors' apparent affinity towards the neurotransmitter. Most of the GRAB sensors have transmitter-insensitive mutant (mut) for comparison with GRAB sensors to check signal specificity. The number represents the version of the sensor. The higher the version, the better the performance.
A2: Sometimes also referred to as DiO (Double-floxed inverse Orientation), FLEx systems are generally considered "Cre-On" meaning that your gene of interest starts in the inverse/antisense "off" position and is flipped to the sense (on) orientation in the presence of Cre. A similar strategy can be employed for "Cre-off" (i.e. your gene of interest starts in the sense orientation and Cre is used to flip it to "off"). These Cre-off switches may be referred to as DO (Double-floxed Orientation), or FDIO, and could be used to study the physiology or behavior of certain cell populations.
A3: GRAB sensors are genetically encoded sensors for neurotransmitters, similar to other genetically encoded sensors (e.g. GCaMP). They could be expressed in specific cell-types and are able to detect dynamics of extracellular neurotransmitters in cultured cells, brain slice and living animal (e.g. fly, zebrafish, mice, rat, monkey etc.). For the aspect of instruments, GRAB sensors could perform well in epifluorescence microscopy, confocal microscopy, 2P microscopy and fiber photometry recording.
A4: Yes. We can record good signals from GRAB sensor after expression for 3 to 6 months in mice brain by AAV infection.
A5: No. The change of fluorescent intensity only indicates the dynamics of corresponding neurotransmitter, which has no direct correlation with excitability of neurons. Another approach is needed for detection of neuron excitability.
A6: The tested GRAB sensors show minimal coupling with G protein and there is no internalization of GRAB sensors induced by coupling with β-arrestin.
A7: The tested GRAB sensors have no or very little specific effect on neurons. For example: 1) Simultaneous patch-clamp recordings showed that the resting membrane potential, input resistance, membrane time constant and average spiking frequency of GACh2.0-expressing CA1 neurons have no significant difference comparing with nearby control non-expressing neurons. Moreover, AMPA, NMDA, and GABAergic responses, as well as paired pulse facilitation of AMPA responses in GACh2.0-expressing neurons, remained unchanged. Moreover, chronic AAV expression of GACh2.0 in the dentate gyrus of hippocampus did not affect the Cal590 fluorescence responses elicited by high K+. We made acute cortical slices after chronic in vivo lentiviral expression of GACh2.0 in barrel cortical L5 pyramidal neurons in mice. Simultaneous patch-clamp recordings showed that GACh2.0-expressing and control non-expressing L5 pyramidal neurons displayed the same resting membrane potential, input resistance, membrane time constant, and average spiking frequency, as well as AMPA, NMDA, and GABAergic responses, and paired pulse facilitation of AMPA responses (Jing et al., 2018, Nature Biotechnology). 2) The experiment on brain slices in hippocampus in mice and on larvae of zebrafish showed that the expression of NE1m had no effect on calcium signal (Feng et al., 2019, Neuron). We co-expressed 5-HT1.0 and jRCaMP1a in fly, and observed no change in calcium signal compared to the non-expressing fly (Wan et al., 2020, bioRxiv).
A8: No. GRAB sensors can be used on cultured cells, tissues, organoid and experimental animals, but not in clinical.
Alford SC, Wu J, Zhao Y, Campbell RE, Knöpfel T. Optogenetic reporters. Biol Cell. 2013 Jan;105(1):14-29. doi: 10.1111/boc.201200054. Epub 2012 Dec 20. PMID: 23126299.
Chefer VI, Thompson AC, Zapata A, and Shippenberg TS (2009). Overview of Brain Microdialysis. Current Protocols in Neuroscience 47, 7.1.1–7.1.28.
Jaquins-Gerstl A, and Michael AC (2015). A review of the effects of FSCV and microdialysis measurements on dopamine release in the surrounding tissue. Analyst 140, 3696–3708.
Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong WH, Folk RW, Broussard GJ, Liang R, Jang MJ, Zhong H, Dombeck D, von Zastrow M, Nimmerjahn A, Gradinaru V, Williams JT, Tian L. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science. 2018 Jun 29;360(6396):eaat4422. doi: 10.1126/science.aat4422. Epub 2018 May 31. PMID: 29853555; PMCID: PMC6287765.
Robinson DL, Venton BJ, Heien ML, and Wightman RM (2003). Detecting subsecond dopamine release with fast-scan cyclic voltammetry in vivo. Clin Chem 49, 1763–1773.
Robinson DL, Hermans A, Seipel AT, Wightman RM. Monitoring rapid chemical communication in the brain. Chem Rev. 2008 Jul;108(7):2554-84. doi: 10.1021/cr068081q. Epub 2008 Jun 25. PMID: 18576692; PMCID: PMC3110685.
Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009 May 8;324(5928):804-7. doi: 10.1126/science.1168683. Erratum in: Science. 2009 Dec 11;326(5959):1482. PMID: 19423828; PMCID: PMC2763207.
Sun F, Zhou J, Dai B, Qian T, Zeng J, Li X, Zhuo Y, Zhang Y, Wang Y, Qian C, Tan K, Feng J, Dong H, Lin D, Cui G, Li Y. Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nat Methods. 2020 Nov;17(11):1156-1166. doi: 10.1038/s41592-020-00981-9. Epub 2020 Oct 21. PMID: 33087905; PMCID: PMC7648260.
 Loading ....
Loading ....
