Solutions for Methylation Library Preparation
Published On 01/23/2025 5:00 PM
DNA methylation—a better tumor early screening marker
Cancer is one of the leading causes of morbidity and death in the world, and about 1 out of every 6 deaths worldwide is due to cancer. According to the "2022 National Cancer Data Report", there are 4.064 million new cases of cancer in the country, and almost 8 people suffer from cancer every minute. Combined with the latest 2020 global cancer burden data previously released by the WHO.The development of cancer is a multi-stage slow process, which generally takes 10 to 15 years from normal to precancerous lesions. Early detection, early intervention, and early treatment are currently the best means of cancer prevention and treatment. At the same time, early detection, early intervention, and early treatment can also greatly improve the effective treatment and survival of patients.
Studies have found that the occurrence of cancer is accompanied by changes in DNA methylation patterns, including retroelements, centromeres, and hypomethylation of proto-oncogenes. Changes in methylation patterns play a key role in the occurrence and development of tumors, and exist almost throughout the development of tumors including precancerous lesions. Therefore, more and more studies will use DNA methylation detection as a marker for early screening of cancer.
At Biohippo, we partner with Yeasen Biotechnologies, a leading provider of molecular diagnostics products, to offer the research tools necessary for detecting DNA methylation.
NO.1 DNA Methylation Bisulfite Kit—— Cat No. BHN20800024
The Hieff Superfast DNA Methylation Bisulfite Kit (Column-based) rapidly converts unmethylated cytosines in DNA samples to uracil, while leaving methylated cytosines unchanged. During high-temperature bisulfite treatment, double-stranded DNA denatures into single strands. In the presence of HSO3-, cytosine residues undergo deamination and are converted into uracil, with methylated cytosines remaining unaltered. In subsequent PCR amplification, uracil is replaced by thymine (T). The conversion process takes only 5 minutes, accommodating DNA input ranging from 100 pg to 2 ug, and achieves a conversion efficiency of >=99% for unmethylated cytosines. The converted DNA is suitable for downstream applications such as PCR amplification and NGS sequencing.
Product Highlights:
- Low input: can meet the conversion of 500 pg-2 μg samples
- High conversion efficiency: conversion rate ≥ 99%
- Easy to operate: conversion can be completed within 3 hours
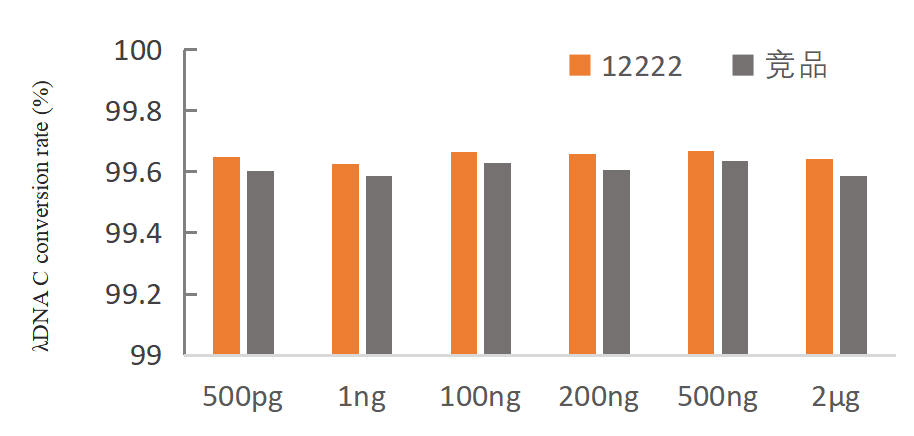
Figure 1. conversion rate of λDNA C in gDNA of 293T cells with different inputs
NO.2 The Hieff NGS dsDNA Methyl Library Prep Kit——BHS20800026
The Hieff NGS dsDNA Methyl Library Prep Kit for Illumina is a double-stranded DNA methylation library preparation kit developed for the Illumina high-throughput sequencing platform. This kit utilizes appropriate methylated adapters for ligation, followed by treatment with bisulfite or enzyme conversion, and then a PCR amplification step to construct methylated libraries for sequencing on the Illumina platform. All components of the kit undergo quality control and functional validation to ensure stable library output, supporting various types of DNA samples including gDNA, cfDNA, and FFPE. The recommended input DNA amount for this kit is 10 ng to 1000 ng, and it is compatible with commonly used conversion protocols.
NO.3 ssDNA Assay Kit——BHE20800020
ssDNA Assay Kit is a single strand DNA (ssDNA) fluorescence quantitative detection kit with good linear relationship between 1-200 ng, which is characterized by high sensitivity.Product Highlights:
- Good linear range: R2>0.99;
- High sensitivity and wide application range: quantification of 1~200 ng samples;
- Strong tolerance: good tolerance to conventional pollutants such as proteins, salts, detergents, etc.
This entry was posted in
Product Literature
,Application and Technique Notes
 Loading ....
Loading ....
